What did motivate the authors to write this review?
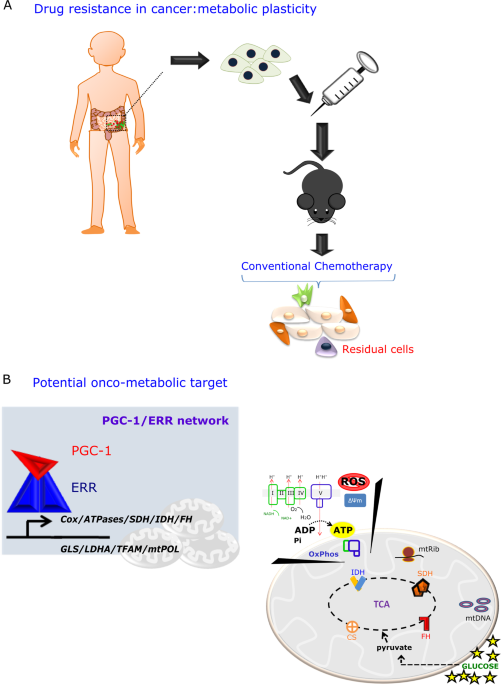
We wanted to understand how to therapeutically explore biological hallmarks acquired during cancer progression and its application in precision oncology. We are motivated by the need to introduce new technologies, such as Cryo-EM. It will highlight the importance of progress that can be fulfilled well beyond long-established technology to answer important questions in cancer metabolism. Particular, I was motivated to study cancer metabolism together with structural biology because once we could enable structural determination for native state complexes that are still not accessible to X-ray crystallographic analysis it might provide new hope for drug discovery and patient treatment against drug resistance in cancer. Furthermore, this knowledge could lead to a better understanding of the specific type of cancer and patients who are responders and who would benefit from the pharmacological targeting of the PGC-1/ERR network.
What is the biggest challenge behind this project?
Cryo-EM technology has several advantages over X-ray crystallography analysis because the protein is frozen in its native state which allows to develop a high-resolution biological structure but, in practice, still many challenges remained, including optimized sample preparation, working on smaller proteins, advances in image processing, advances in detectors, advances in transmission EM, automated data collection systems, automated model building, and structure refinement. The PGC-1/ERR complex is considered a smaller protein complex with a high structural flexibility. The biggest challenge behind this project is the sample preparation step from a small and flexible protein complex, which is bringing more challenge to the table. However, extensive research in our Institute is pushing the project to the next level. Currently, we do have couple of evidences showing that this small complex interacts with an important target in signal transduction cascades that controls oxidative mitochondrial metabolism (OxPhos) and cancer cell survival.
What next?
In our study, we wanted to cover various aspect of cancer metabolism using the power of Cryo-EM technology. We expect to deeper mechanistic understanding of signaling cascades involved in cancer therapeutic resistance. We hope that structural biology would encourage others to develop similar studies and, in the near future, it would offer of developing new treatments targeting metabolic-addictive cancer cells.
Humberto De Vitto
Postdoctoral Researcher, The Hormel Institute, University of Minnesota




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in