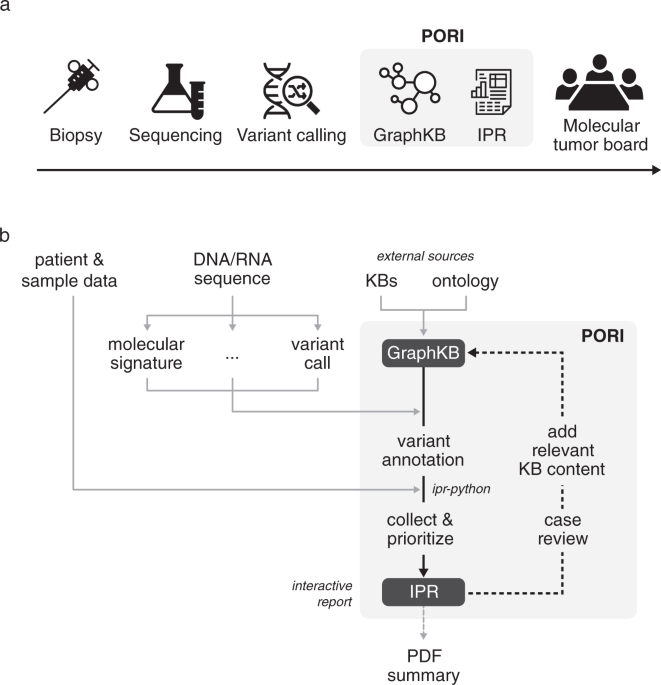
One of the most significant challenges in precision medicine is interpreting and communicating an expanding domain of clinically relevant molecular biomarkers. The Platform for Oncogenomic Reporting and Interpretation (PORI, https://bcgsc.github.io/pori) described in our recent publication Nature Communications simplifies this process by integrating multiple knowledge bases and ontologies into one platform. PORI is composed of the graph-based knowledge base, GraphKB, and reporting software, Integrated Pipeline Reports (IPR) and is the first open-source, production-ready and scalable software solution. PORI accommodates variant, gene expression and complex molecular data and reports clinically relevant knowledge curated from multiple external knowledge resources into a genomic report that can be disseminated to clinical and research experts.
The Origin Story
The Platform for Oncogenomic Reporting and Interpretation (PORI) began as a way to support the analysis of individual cases in the Personalized OncoGenomics (POG) program (NCT02155621). From sequencing the first patient in 2012, POG was one of the first clinical trials to biopsy and sequence tumour whole genomes and transcriptomes from patients with advanced cancer, integrating these findings into real-time treatment planning. A lack of existing informatics tools needed to fully integrate whole genome and transcriptome data and the growing domain knowledge required us to start from scratch in building many of the methods, tools, and standards for the POG program. Throughout the course of this study, the manual time required to review variants and the pertinent literature has been the most labour-intensive analysis step of the process.
The platform's initial development began, as it often does in software, with the tedium of a difficult manual task we wanted to improve. Analysts would routinely spend more than a week searching the literature and looking up variants and their clinical significance in several online databases for each sequenced tumour sample. To save time in data collection, the genome analysts and developers at the Genome Sciences Centre (https://bcgsc.ca) began to store this information in an early version of what later became GraphKB. GraphKB began as every biologist's favourite tool, a simple spreadsheet. Future iterations involved a proper database, but it is difficult to represent ambiguity and hierarchical relationships in a traditional (relational) database, so we quickly moved on to what has become the current implementation of GraphKB using a graph model. GraphKB stores information from literature and external databases in such a way that we can match variants found in a patient's tumour sample to the knowledge base in an automated manner, limiting the potential for manual mistakes.
One of the many difficulties we had to overcome with POG was translating knowledge between the bioinformatics and research scientists to the clinicians and clinical nurses. After data is analyzed and reported from our bioinformatics pipeline, each case is presented to a multi-disciplinary group of clinicians, pathologists, and research scientists known as a molecular tumour board for discussion. The diverse nature of the molecular tumour board allows us to have a vast range of expertise to pull from, but it also means that every week, everyone has to be ready to learn about niche topics and rare diseases or genes they may never have encountered before. This is why we developed the reporting component of PORI, Integrated Pipeline Reports (IPR), with the involvement of the clinicians and clinical nurses. We conducted semi-regular meetings to discuss updates and propose changes to the report (Figure 1).

Even with a well-designed report, there is a significant learning curve for everyone involved, so the interactive nature of the report allows us to make this easier by providing explanatory material and links alongside the content of the report. One of the major outcomes of these design meetings was the need for both a condensed and extended version of the report. To accommodate the busy schedules of our report users, we have a report overview on the main page which allows users to quickly see the main points of the report. If there is a particularly interesting or actionable variant the user needs to have the full information on, more detail is made available in subsequent sections.
The Importance of open source
Open source and open data are incredibly important to supporting reproducible science as well as enabling critical review of scientific procedures. Initiatives that engage and leverage the expertise of the scientific community and provide open source resources such as CIVIC (https://civicdb.org) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) are fundamental for the freely available access to clinically relevant information to the wider clinical and scientific community. As we’ve been very fortunate to leverage tools from the open source science and software community we are fully committed to giving back with our own contributions. A significant challenge to open-sourcing this platform was the sheer amount of code involved. Through the release of PORI we have open sourced our entire reporting platform consisting of more than 10 code repositories worked on by more than a dozen developers.
Participation in several collaborations across multiple sequencing centres has highlighted the importance of standardizing genomic data analysis and reporting. Many institutions are using private custom, or proprietary solutions which makes this a difficult task. This was a large part of the motivation for open sourcing our own reporting platform. We are excited to continue this platform for both our own needs but also to support the rest of the precision oncology community.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in