Acute myeloid leukaemia (AML) is characterized by recurrent genetic aberrations in more than 90% of cases1, however, only some have a prognostic value and persistent mutations might therefore reflect a re-established premalignant state or truly active disease causing relapse2. For instance, certain persistent mutations in genes such as DNMT3A, TET2 or ASXL1 (DTA) were not associated with a worse outcome3 and are also the most prevalent gene mutations defining age related clonal hematopoiesis (ARCH2,4,5) or clonal hematopoiesis of indeterminate potential (CHIP6). The increasing availability and sensitivity of NGS applications have driven attempts to further identify molecular markers for minimal residual disease (MRD) detection. We aimed to dissect the nature of co-mutations in NPM1 mutated AML where the detection of NPM1 transcripts allows highly specific and sensitive detection of complete molecular remission (CMR).
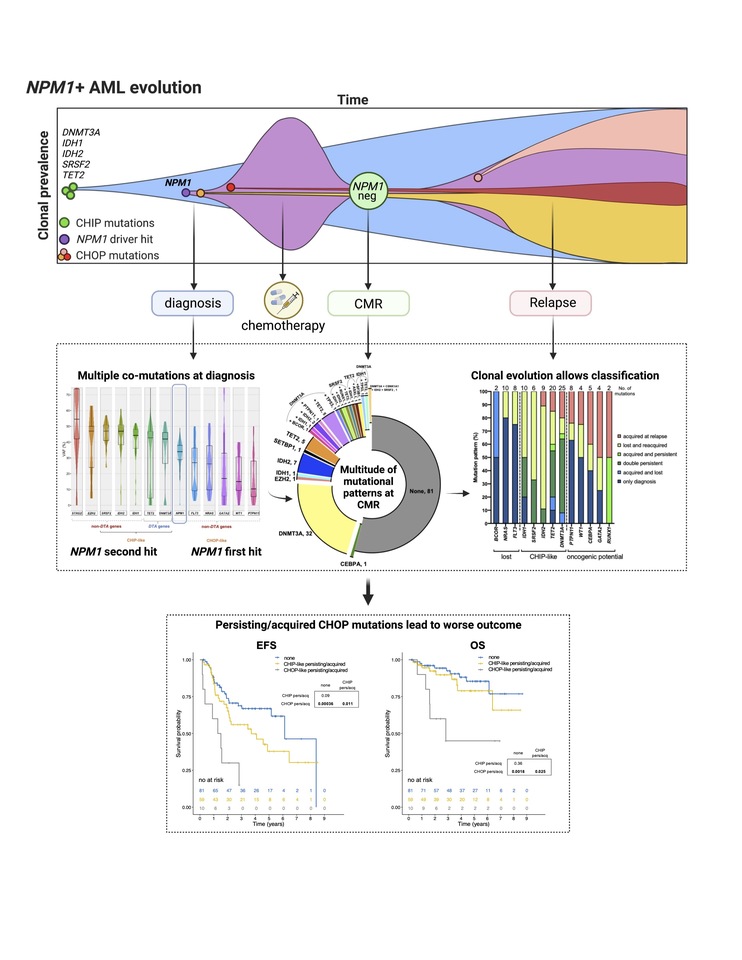
We analysed 150 consecutive NPM1 mutated AML patients who achieved CMR following intensive treatment by next generation sequencing on paired samples at diagnosis, CMR and relapse (38/150 patients). We identified the persistence of non DTA-mutations at CMR in a significant proportion of patients (15%), and expand previous studies showing that also in NPM1 mutated AML the persistence of non-DTA-mutations in remission is detrimental3,2,7 (EFS HR=2.7, p=0.003; OS HR=3.6, p=0.012). For this entity, MRD detection in clinical remission has long been established and is more informative for survival than the detection of co-mutations such as FLT3-ITD or DNMT3A.8,9 Our data on NPM1mut AML in CMR suggest that the persistence of non-DTA mutations represents molecular residual disease. Furthermore, we show for the first time that also the acquisition of non-DTA mutations at CMR is an adverse prognostic factor in NPM1mut AML.
Based on clonal evolution analysis of diagnostic, CMR and relapse samples, we redefine pre-malignant mutations and include IDH1, IDH2 and SRSF2 with the DTA genes in this newly defined group (CHIP-like mutations). This was further justified by the fact that also persistent/acquired IDH1/2 or SRSF2 mutations had no impact on survival along with mutations in DNMT3A and TET2. We defined all other mutations (i.e. FLT3-TKD, GATA2, NRAS, PTPN11, WT1, TP53, RUNX1) as CHOP-like (clonal hematopoiesis of oncogenic potential) mutations. Only the persistence or acquisition of CHOP-like mutations was significantly associated with an inferior outcome (EFS HR=4.5, p=0.0002; OS HR=5.5, p=0.002). By multivariate analysis, incorporating known risk factors such as allogeneic stem-cell transplantation, aberrant karyotype, gender and age, we found that the persistency/acquisition of CHOP-like mutations at CMR was an independent predictor of outcome (HR=7, 1.6-30, p=0.009), and was stronger compared to the classification of DTA mutations alone.
Moreover, CHOP-like mutations were detrimental even when detected at relapse (HR=4.5, p=0.01). On the other hand, the detection of either CHIP or CHOP mutations at diagnosis did not have any impact on OS, highlighting the importance of sampling at CMR or relapse.
In conclusion our data allows the distinction of molecular residual disease from the persistence of a pre-malignant state which likely does not affect prognosis. We propose a model where CHIP-like mutations define a pre-malignant state in NPM1mut AML which require the additional acquisition of driver mutations. While CHOP-like mutations might drive disease progression on their own.
CMR in NPM1mut AML is an independent factor for good risk disease8, however up to 30% of patients with CMR relapse. Here we provide a clinical tool where the detection of oncogenic mutations at CMR, is an independent prognostic factor facilitating early intervention in those patients. Studies with MRD guided therapy in AML show promising results: in the RELAZA2 trial, patients with MRD positive AML following conventional chemotherapy or allogeneic transplant were treated with azacytidine and showed a clinical meaningful benefit.13 Based on the QUAZAR trial an oral formulation of azacytidine (CC-486) is the first approved maintenance therapy for AML14, which could be especially worthy in patients with persistent CHOP-like mutations.
Patients relapsing with persistent or novel CHOP-like mutations represent an unmet clinical need and strategies like the RELAZA protocol, maintenance with demethylating substances13,15 or treatment with novel agents16 could improve outcome.
In summary, our data show that even in the relatively favorable context of NPM1mut AML following CMR, modern NGS based screening can identify patients at risk in order to develop personalized therapeutic strategies aimed at eradicating MRD and molecular residual disease to prevent relapse. The conduction of NGS-based MRD-guided clinical trials dedicated to this subset of NPM1mut AML patients is highly warranted.
For further details please read our paper.

Figure | In NPM1 mutated AML, the detection of CHOP like mutations at molecular remission confers inferior outcome. Upper box: schematic representation of clonal prevalence over time in AML. CHIP mutations often arise early in leukemia development and in few cases are followed by driver hits such as NPM1 which lead to overt AML. Patients with NPM1 mutated AML are usually approached with intensive chemotherapy and often reach complete molecular remission (CMR), defined by the absence of NPM1 transcripts. Surviving clones bearing oncogenic (CHOP) mutations may be selected and eventually expand leading to clinical relapse. Middle box: by deep sequencing we analyzed paired samples of 150 NPM1 mutated AML patients at diagnosis, CMR and when available, relapse (n=38). Multiple co-mutations are detectable at diagnosis with either smaller or larger clonal abundance than NPM1. At CMR, while NPM1 is undetectable, a multitude of mutations can persist. Taking advantage of longitudinal sampling, we performed a clonal evolution analysis and confirmed the presence of mutations with an indetermined role in AML pathogenesis, potentially related to early stages of leukemogenesis (CHIP), vs mutations with a clear oncogenic potential (CHOP). Bottom box: survival analysis shows that the detection of CHOP mutations at CMR confers inferior prognosis in NPM1 mutated AML patients.
1. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209-2221.
2. Rothenberg-Thurley M, Amler S, Goerlich D, et al. Persistence of pre-leukemic clones during first remission and risk of relapse in acute myeloid leukemia. Leukemia. 2018;32(7):1598-1608.
3. Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N Engl J Med. 2018;378(13):1189-1199.
4. Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498.
5. Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487.
6. Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16.
7. Morita K, Kantarjian HM, Wang F, et al. Clearance of Somatic Mutations at Remission and the Risk of Relapse in Acute Myeloid Leukemia. J Clin Oncol. 2018;36(18):1788-1797.
8. Ivey A, Hills RK, Simpson MA, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med. 2016;374(5):422-433.
9. Schnittger S, Kern W, Tschulik C, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114(11):2220-2231.
10. Patel JL, Schumacher JA, Frizzell K, et al. Coexisting and cooperating mutations in NPM1-mutated acute myeloid leukemia. Leuk Res. 2017;56:7-12.
11. Cappelli LV, Meggendorfer M, Dicker F, et al. DNMT3A mutations are over-represented in young adults with NPM1 mutated AML and prompt a distinct co-mutational pattern. Leukemia. 2019;33(11):2741-2746.
12. Hollein A, Meggendorfer M, Dicker F, et al. NPM1 mutated AML can relapse with wild-type NPM1: persistent clonal hematopoiesis can drive relapse. Blood Adv. 2018;2(22):3118-3125.
13. Platzbecker U, Middeke JM, Sockel K, et al. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multicentre, phase 2 trial. Lancet Oncol. 2018;19(12):1668-1679.
14. Wei AH, Döhner H, Pocock C, et al. Oral Azacitidine Maintenance Therapy for Acute Myeloid Leukemia in First Remission. N Engl J Med. 2020;383(26):2526-2537.
15. Huls G, Chitu DA, Havelange V, et al. Azacitidine maintenance after intensive chemotherapy improves DFS in older AML patients. Blood. 2019;133(13):1457-1464.
16. Cortes JE, Heidel FH, Hellmann A, et al. Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia. 2019;33(2):379-389.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in