Invasive cervical cancer kills nearly 311,000 women each year worldwide with an estimated 50% increase in deaths by 2040. Women with advanced or recurrent cervical cancer often develop resistance to current chemotherapy options, and >90% die within 2 years. Thus, identification and characterization of novel, clinically actionable target genes remains a crucial avenue of research for development of more effective therapeutic approaches for women with cervical cancer. Our recent study published in the British Journal of Cancer describes a workflow for identification and prioritization of putative cervical cancer target genes from tumor HPV integration signatures.
How it started
I had just finished my second post-doc studying the effects of estrogen on hippocampal memory (neuroscience was my first love!) when I began my tenure in Dr. Janet Rader’s lab at the Medical College of Wisconsin in 2013. I was immediately immersed into the world of cervical cancer genomics, helping collect and process cervical tumor samples for something called The Cancer Genome Atlas (TCGA). Little did I know we would still be publishing on these samples some 8 years later…
In addition to processing cervical cancer specimens for TCGA, our lab had also just begun a large HPV sequencing project aimed at identifying HPV integration events in a cohort of invasive cervical cancer (ICC) samples curated and homed by our lab. One of the most intriguing findings from this study was identification of multiple HPV integration events in the long noncoding RNA (lncRNA), PVT1. I thought to myself, “Why integrate here? What is special about this gene/gene region?”. Not much was known about PVT1 at the time, so we decided to take a closer look into how the lncRNA may function in cervical cancer. Cut to 2016 and we are publishing the first paper supporting an oncogenic role for PVT1 in cervical cancer. So, this is where the original idea behind our newly published paper was born – using HPV integration as a sort of flashlight to illuminate novel cervical cancer-promoting genes.
How it's going
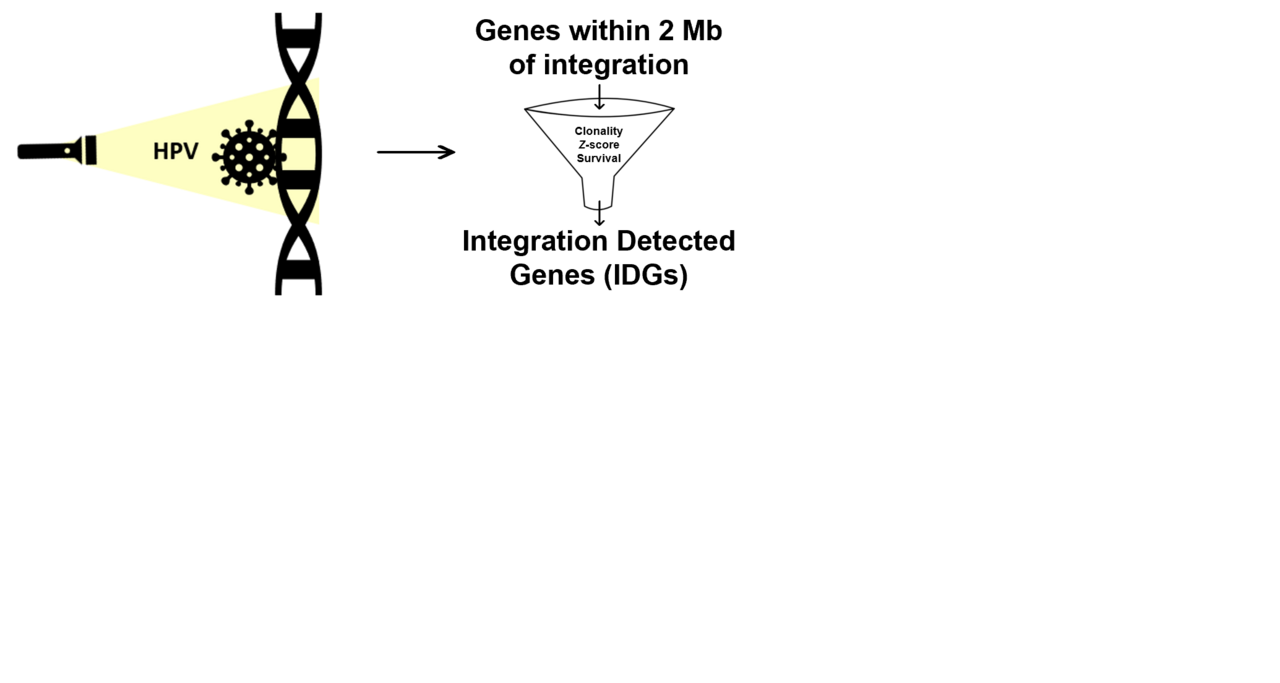
We decided to return to some of our TCGA samples (MCW-TCGA), using our new flashlight for pinpointing putative cervical cancer target genes - or, as we like to call them, integration detected genes (IDGs). We leveraged the large amount of existing multi-omics TCGA data and used residual, matched tumor sample for PacBio long-read sequencing for thorough functional/structural annotation of HPV integration events. We began with 8 MCW-TCGA samples, finding 267 unique HPV-human breakpoints comprising a total of 87 HPV integration events.
Next, we recorded all genes within 2 Mb on either side of each integration event giving us our initial list of candidate IDGs. Candidate IDGs were then prioritized via a filtering system based on the following criteria: 1) clonal representation of integration event, 2) integrated tumor-specific expression (Z-score), and 3) association with cervical cancer survival. From the 8 samples examined, our filtering system successfully cut the number of candidate IDGs from 3,134 to 84. Four of the final 84 IDGs were then chosen for in vitro functional assays employing siRNA-mediated knockdown to examine their potential roles in cervical cancer cell proliferation, migration, and colony formation.
Big takeaway and what's next
Although our pipeline for identifying and filtering candidate IDGs worked reasonably well, we learned a lot and continue to explore ways in which it can be improved. We know that we cannot count on all integration events being associated with clinically relevant targets. We also know that the "chicken and egg" dilemma still exists regarding the role of HPV integration in cervical carcinogenesis: are HPV integration-induced alterations to the human genome a critical driver of cervical cancer progression or were the host genomic alterations already present in the tumor prior to viral integration? Luckily for our purposes, it may not really matter.
What we do know is that our results strongly reinforce the idea that HPV integration frequently disrupts important cancer genes, providing further support for use of in-depth integration analysis as a mechanism for pinpointing novel drivers of cervical cancer. Our future work will continue implementing our pipeline in this and other large cervical cancer cohorts so that we may fine-tune the filters for better IDG prioritization. We will also continue to examine candidate IDGs highlighted in the current study to further determine their specific function and clinical potential in cervical cancer. Finally, as we apply our pipeline to more and more tumor samples, is it our hope that similarities across tumors/integrations will begin to reveal themselves so that they may be used for improved stratification of cervical cancer patients into clinically actionable cohorts.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in