Lung cancer is the leading cause of cancer-related deaths worldwide with annually 2.1 million new lung cancer cases and 1.8 million deaths1. Over the past 30 years, the overall five-year survival for lung cancer in the United States has increased from 12% to over 19% and continues to rise2, which is attributable to early detection screening programs and targeted therapies.
Lung adenocarcinoma is the most common histological subtype of lung cancer. Early-stage LUAD patients have better prognosis than patients with advanced stage disease; however, about 30-50% of early-stage patients develop metastasis after surgical resection. Overall 5-year survival rate for stage 1 and 2 non-small cell lung cancer patients is 70% and 35%, respectively3,4. Lung adenocarcinoma is a histologically heterogeneous disease, and is associated with diverse tumor invasion phenotype and clinical outcomes5.
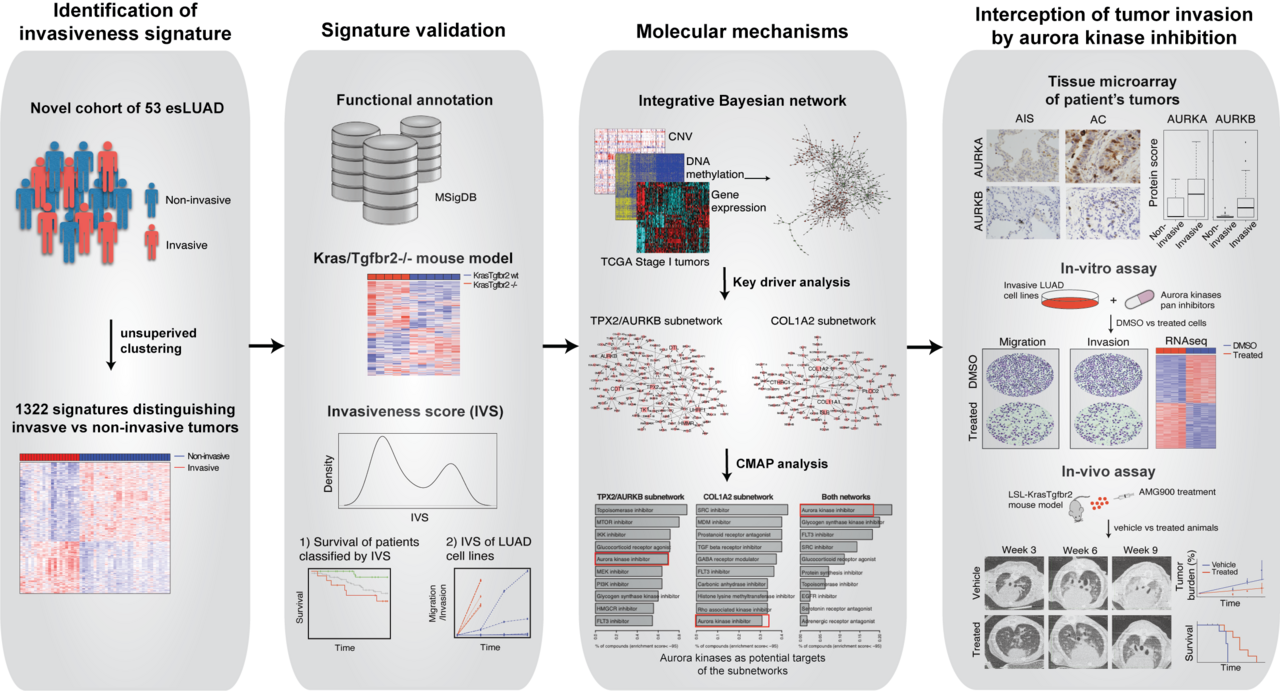
Invasiveness is one of cancer hallmarks and is directly related with metastatic potential and clinical outcomes of the tumor6. In this study, we present a genomic approach that that targets molecular subtypes of early-stage lung adenocarcinoma (esLUAD) and therapeutic promise for this important patient group.
In this study we performed molecular profiling of a cohort of 53 histologically heterogeneous esLUAD tumor samples. An unsupervised hierarchical clustering resulted in two distinct groups consistent with their histological subtypes. Group 1 was enriched for pathological aggressive subtypes acinar, micro papillary, papillary and solid while Group 2 consisted of less invasive or non-invasive tumors such as MIA, AIS, and lepidic predominant and hence annotated as “Invasive” and “Non-invasive” group, respectively. Together, 1,322 DEGs that distinguished the Invasive and Non-invasive tumors were annotated as “invasiveness signature genes”. The invasiveness signature genes were separated into 526 genes up-regulated in the Invasive group (pro-invasive signature genes) and 796 genes up-regulated in the Non-invasive group (indolence signature genes), where pro-invasive signature genes showed enrichment of EMT and angiogenesis pathways which are major hallmarks of tumor invasion and metastasis functions6.
We hypothesized that our gene signatures could predict survival of esLUAD patients. Using the pro-invasive and indolence signature genes, we developed an Invasiveness Score (IVS) based on elastic net to estimate tumor’s invasiveness for seven independent lung adenocarcinoma cohorts for stage I and II tumors. In all 7 independent datasets “Invasive” groups had significantly inferior survival compared to “Indolent” groups.
Next, to validate the biological implications of the IVS-based ranking, we experimentally assessed cell invasiveness using in vitro migration and invasion assays. We selected 8 high IVS with 5 KRAS mutant: H1373, SK-LU-1, H1792, A549, and H2009; 3 KRAS wild type: H1755, H1650 and H1975; and 5 low IVS HCC78, H3255, Calu3, HCC1833, HCC2279 cell lines. All 8 high IVS cell lines showed higher transwell migration and invasion within 48hr time period independent of their KRAS status compared to low IVS score cell lines, suggesting that the invasiveness phenotype predicted by our IVS-based ranking is significantly associated with experimentally observed biological phenotype.
The biological mechanism that drives the transformation of tumors from indolence to invasiveness is not well characterized. To that direction, we constructed a molecular regulatory network for esLUAD by integrating gene expression, CNV, and methylation profiles of the stage I LUAD samples in TCGA using the software package RIMBANET 7,8. The pro-invasive signature genes formed two large closed subnetworks. The first subnetwork was regulated by TPX2, AURKB, and 5 other key regulators (referred as TPX2/AURKB subnetwork) and the second subnetwork was driven by collagen associated genes COL1A2 and COL11A1, and 3 other key regulators (referred as COL1A2 subnetwork). To identify drugs that could impact tumor invasiveness, we searched the Connectivity map (CMAP) database9 to identify small molecules that could perturb the two pro-invasive subnetworks. Aurora kinase inhibitor was the strongest perturbagen class to reverse the expression of the genes in the both subnetworks. Taken together, the genomic analyses suggest that aurora kinases represent potential regulators of invasiveness that are vulnerable to small molecular inhibitors.
Aurora kinase A and B are aberrantly expressed in several tumor types10-15, and promote cancer proliferation and survival owing to their functions in cell cycle and mitosis16. However, their role in regulating the invasiveness and migratory phenotypes in human tumors is not well characterized. We used genetic and small molecule inhibition approaches to examine the effect of AurA and/or AurB on tumor cell migration and invasion. We performed CRISPR/Cas9 deletion of AURKA and AURKB in highly invasive H1792 and A549 cells. Repressing AURKA or AURKB alone did not alter migration and invasion in either cell line but, deleting both AURKA and AURKB together demonstrated significant reduction in migration by 60% and 50% and invasion by 70% and 60% in H1792 and A549 cells, respectively at 48hr, adjusting for cell number. We performed small molecule inhibitor studies using pan-aurora kinase inhibitors, AMG900 and PF-03814735. Both pan-aurora kinase inhibitors significantly suppressed transwell migration, transwell invasion and wound healing ability for LUAD cell lines. These results suggest that AurA and AurB have redundant functions in regulating lung tumor invasiveness and pan-inhibition is required to impact tumor cell invasion.
To comprehensively understand the molecular mechanisms driven by inhibition of aurora kinases, we examined gene expression profiles of A549 and H1792 cells treated with an aurora kinase inhibitor (AMG900). Analysis of DEGs on treatment with inhibitor, showed enriched of MTORC1_SIGNALING in both LUAD cells. Consistent with the RNAseq transcriptomic analysis, we observed decreased p-AKT (Thr308) and p-mTOR (Ser2448) protein expression as well as suppressed expression of p-S6-kinase (Ser371) and p-4E-BP1 (Thr37/46) downstream targets of mTOR/Akt pathway. The lung adenocarcinoma pro-invasive signature was enriched for EMT signature genes. We observed down-regulated mesenchymal markers N-Cadherin and Vimentin and up-regulated E- Cadherin in A549 and H1792, suggesting that aurora kinases mediate invasiveness and cell motility by regulating MTOR/AKT and EMT pathways.
To investigate the pre-clinical utility of aurora kinase inhibitors we treated our transgenic invasive LUAD mouse model LSL-Kras(G12D)Tgfbr2-/- with AMG900. We previously showed that loss of Tgfbr2 in Kras G12D mutant condition induces highly invasive phenotype17 in the transgenic mouse model. Micro-CT images demonstrated significantly lower tumor density in AMG900 treated animals. Histopathological analysis identified several invasive and in situ tumors with bulky nodules. While solid and acinar patterns increased in control animal lungs, lungs from AMG900 treated animals showed suppression of invasiveness in mouse lungs. Histopathological analysis of lung tumors showed a reduction in invasive tumor lesions with aurora kinase inhibition, adjusted for tumor size, further supporting a direct link between aurora kinase inhibition and tumor invasion phenotype. We evaluated the impact of aurora kinase inhibition on the tumor microenvironment composition. AMG900 treatment led to significant reduction in tumor infiltrating CD31+ neovessels and collagen deposition in AMG900 treated group as compared to vehicle treated group. While, vehicle group animals died within 18 weeks of treatment, AMG900 group survived till 35 weeks of treatment. Taken together, our in vivo transgenic data indicated that AMG900 treatment intercepts progression of invasive lung adenocarcinoma and leads to better survival in this animal model of early-stage invasive lung adenocarcinoma.
Insights of the study
In advanced lung cancer, genomic dissection of clinical subtypes has resulted in identification of actionable alterations leading to development of targeted therapeutics and increased overall survival. Our premise is that similar approaches applied to early-stage lung cancer could be of great clinical impact. We show the clinical and biological significance of lung adenocarcinoma invasiveness as a key driver of tumor progression, metastasis, and clinical outcome in early-stage disease. In this study, we generated gene signatures of lung adenocarcinoma invasion and indolence and validated the biological and clinical importance of these signatures in independent cohorts, human tissues, human cell lines, and in genetically engineered mouse models of invasive lung adenocarcinoma. We mined genomic data acquired from early-stage human lung adenocarcinoma specimens to identify signatures of tumor invasiveness and we used network-modeling approaches to identify key drivers susceptible to small molecule inhibition in vitro and in vivo. Our preclinical studies exhibit abrogation of progression and spread of invasive LUAD tumor with AMG900 treatment thus illustrating the importance of aurora kinase inhibition as powerful therapeutic opportunity in clinic.

References:
- Bray, F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394-424 (2018).
- Howlader, N., et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med 383, 640-649 (2020).
- Borczuk, A.C., Kim, H.K., Yegen, H.A., Friedman, R.A. & Powell, C.A. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med 172, 729-737 (2005).
- Uramoto, H. & Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res 3, 242-249 (2014).
- Travis, W.D., et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6, 244-285 (2011).
- Hanahan, D. & Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 144, 646-674 (2011).
- Zhu, J., et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet 40, 854-861 (2008).
- Zhu, J., et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol 10, e1001301 (2012).
- Subramanian, A., et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437-1452 e1417 (2017).
- Bischoff, J.R., et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J 17, 3052-3065 (1998).
- Sen, S., Zhou, H. & White, R.A. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene 14, 2195-2200 (1997).
- Tanaka, T., et al. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res 59, 2041-2044 (1999).
- Lin, Z.Z., et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer 10, 461 (2010).
- Vischioni, B., Oudejans, J.J., Vos, W., Rodriguez, J.A. & Giaccone, G. Frequent overexpression of aurora B kinase, a novel drug target, in non-small cell lung carcinoma patients. Mol Cancer Ther 5, 2905-2913 (2006).
- Qi, G., et al. Aurora-B expression and its correlation with cell proliferation and metastasis in oral cancer. Virchows Arch 450, 297-302 (2007).
- Willems, E., et al. The functional diversity of Aurora kinases: a comprehensive review. Cell Div 13, 7 (2018).
- Borczuk, A.C., et al. Progression of human bronchioloalveolar carcinoma to invasive adenocarcinoma is modeled in a transgenic mouse model of K-ras-induced lung cancer by loss of the TGF-beta type II receptor. Cancer Res 71, 6665-6675 (2011).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in