Post-transcriptional regulation is a rapid and efficient process in which the integrity, diversity, and abundance of a cell’s protein products are adjusted to a changing environment. RNA binding proteins (RBPs) are the master regulators of this process, as they have the ability to bind and regulate thousands of transcripts to modulate gene expression. While most bind to specific mRNA sequences or structures, some associate with mRNA in a highly flexible manner through their low complexity, intrinsically disordered regions (IDR)1,2. Interestingly, some RBP functions extend to transcription factors, in which they associate with chromatin and regulate transcription3.
Dysregulated RBPs have been found in numerous cancers, with several identified in hematopoietic malignancies4,5. For example, MSI2 has been found to drive aggressive myeloid leukemia6. Acute myeloid leukemia (AML) is a hematological malignancy characterized by the uncontrolled expansion of poorly differentiated myeloid cells. The overall survival in adult AML has only improved marginally in the last decades7. To discover novel RBP regulatory pathways in leukemia, we had previously performed an in vivopooled shRNA screen for MSI2’s protein interactors and associated genes. From this screen, the RBP RBMX emerged as a promising oncogenic candidate.
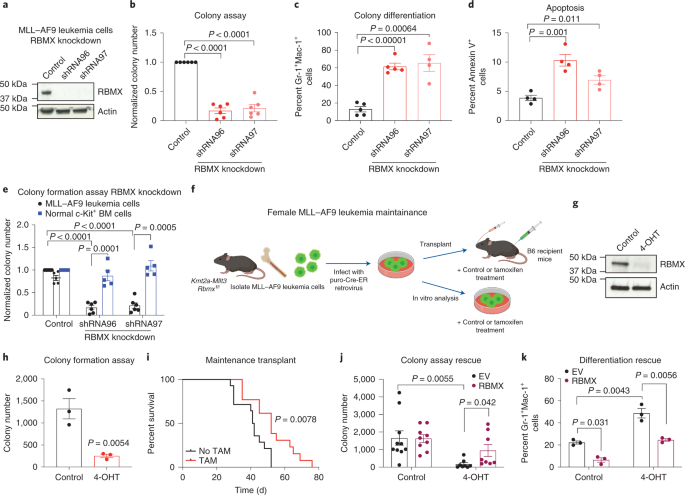
RBMX has been implicated as a tumor suppressor in multiple solid cancers8-11, but its role in blood cancers remains unknown. Therefore, our initial identification of RBMX as a potential oncogenic regulator of leukemia needed to be further evaluated. Using RNA interference approach and conditional knockout murine models, we show that RBMX depletion is detrimental to the survival of MLL-AF9 driven leukemia cells in vitro and inhibits their ability to maintain the disease in vivo. However, we found that Rbmx knockout did not affect the leukemia initiating capacity of this fusion oncogene and RBMX protein was unexpectedly retained in the moribund knockout animals. We later determined that the retained protein band was the retrogene RBMXL1, which shares 95% polypeptide homology with RBMX. RBMXL1 depletion in Rbmx deficient leukemia cells resulted in further delayed leukemogenesis, indicating that RBMXL1 is functionally redundant to RBMX and both genes are required for leukemia development and maintenance. Interestingly, while mRNA levels of Rbmxl1 were similar in the presence or absence of Rbmx, its protein abundance was increased over a long period of Rbmx loss, indicating that it was translationally/post-translationally upregulated. Although the exact mechanism underlying this regulation remains to be elucidated, our study uncovers a genetic compensation by a retrogene in leukemia.
RBMX and RBMXL1 are overexpressed in AML primary patients compared to healthy individuals. Loss of both proteins inhibited cell growth, induced differentiation and apoptosis, and delayed leukemia development in vivoin human AML cell lines and patient samples. In contrast, RBMX and RBMXL1 knockdown in human CD34+stem and progenitor cells reduced colony formation but had no effect on apoptosis. Taken together, our data suggest that RBMX and RBMXL1 are differentially required in leukemia cells versus normal cells in both human and mouse systems.
Mechanistically, RBMX has previously been shown to play pleiotropic roles, including roles in splicing, chromosome segregation, DNA damage response, and maintaining genome integrity via interaction with long-noncoding RNA12-18. However, we found modest splicing defects, no consistent DNA damage as measured by yH2AX signal (unpublished data), nor chromosome segregation defects upon RBMX and L1 knockdown in leukemia cells.
In contrast, our complex karyotyping and ATAC-seq analysis identified a global redistribution of chromatin compaction and an increase in metaphases with breaks and gaps. Using RNA-seq and PAR-CLIP analyses, we revealed that RBMX and RBMXL1 directly bind to mRNAs, reducing transcription of multiple loci, including CBX5 (also known as HP1a - a key component of heterochromatin). Although RBMX and RBMXL1 interact with the 3’UTR and intron 1 of CBX5 mRNAs, we found no effect on CBX5 mRNA stability. Instead, our nascent RNA capture and single molecule FISH determined that RBMX and RBMXL1 control the nascent transcription of the CBX5 locus, via direct interaction with the intron 1 of CBX5 transcripts. This begs the question how an RBP can bind to mRNA targets and regulate transcriptional activity.
The clue might lie in our finding that the IDR but not the RNA-recognition motif (RRM) domains of RBMX mediated the control of CBX5 nascent transcription. Intriguingly, RBMX has recently been shown to directly bind RNA polymerase II and m6A-methylated RNAs through its IDR to regulate alternative splicing in non-blood cells. Furthermore, RBMX interacts with YTHDC1, another m6A reader, and co-regulates the splicing of their common targets in this context14. Remarkably, in our recently published study, we show that YTHDC1 forms a novel type of cellular body, called nuclear YTHDC1-m6A condensates or nYACs, via its IDR to regulate survival of human myeloid leukemia cells. Similarly to RBMX and RBMXL1, loss of YTHDC1 altered expression of its mRNA targets without affecting splicing in leukemia19. Thus, we speculate that (1) these RBPs interact with each other in nYACs, but the function of these bodies is highly cell-context dependent, and (2) they co-regulate transcriptional activity of a subset of m6A-methylated targets in leukemia cells. Our future studies will explore the relationship between these nuclear RNA regulators.
In summary, we uncover that RBMX and RBMXL1 control leukemia cell survival by regulating the chromatin state through their downstream target CBX5, presenting novel therapeutic targets in myeloid malignancies. We show unexpected genetic compensation by a retrogene in leukemia and reveal a new phenomenon for RBPs in regulating transcriptional activity. Our study also provides evidence for the cell-context dependency of RBP functions, with RBPs having opposite roles and tissue/cancer-specific molecular mechanisms of action.
References:
- Nott, T.J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57, 936-947 (2015).
- Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123-33 (2015).
- Xiao, R. et al. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell 178, 107-121 e18 (2019).
- Prieto, C. & Kharas, M.G. RNA Regulators in Leukemia and Lymphoma. Cold Spring Harb Perspect Med 10(2020).
- Pereira, B., Billaud, M. & Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 3, 506-528 (2017).
- Kharas, M.G. & Lengner, C.J. Stem Cells, Cancer, and MUSASHI in Blood and Guts. Trends Cancer 3, 347-356 (2017).
- Maynadie, M. et al. Twenty-five years of epidemiological recording on myeloid malignancies: data from the specialized registry of hematologic malignancies of Cote d'Or (Burgundy, France). Haematologica96, 55-61 (2011).
- Shin, K.H., Kang, M.K., Kim, R.H., Christensen, R. & Park, N.H. Heterogeneous nuclear ribonucleoprotein G shows tumor suppressive effect against oral squamous cell carcinoma cells. Clin Cancer Res 12, 3222-8 (2006).
- Martinez-Arribas, F. et al. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem 97, 1275-82 (2006).
- Ouyang, Y.Q. et al. Expression levels of hnRNP G and hTra2-beta1 correlate with opposite outcomes in endometrial cancer biology. Int J Cancer 128, 2010-9 (2011).
- Hirschfeld, M. et al. HNRNP G and HTRA2-BETA1 regulate estrogen receptor alpha expression with potential impact on endometrial cancer. BMC Cancer 15, 86 (2015).
- Adamson, B., Smogorzewska, A., Sigoillot, F.D., King, R.W. & Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14, 318-28 (2012).
- Munschauer, M. et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 561, 132-136 (2018).
- Zhou, K.I. et al. Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Mol Cell 76, 70-81 e9 (2019).
- Becker, J.S. et al. Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes. Mol Cell 68, 1023-1037 e15 (2017).
- Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45, 6051-6063 (2017).
- Heinrich, B. et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem 284, 14303-15 (2009).
- Matsunaga, S. et al. RBMX: a regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep 1, 299-308 (2012).
- Cheng, Y. et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell (2021).

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in