For the first time in 40 years, new therapies have entered the clinic for the treatment of the most aggressive and lethal form of leukaemia - acute myeloid leukaemia (AML) (reviewed1). Nevertheless, disease recurrence is still common, highlighting the need to identify new drug targets that confer a more durable response. The recently updated European Leukemia Net (ELN) recommendations for diagnosis and management of adult AML,2 highlights genomics-based advancements employed in the development of treatment strategies that may complement or supersede current regimens. However, little consideration is given to the myriad of non-genomic, mitogenic and environmental contributors to sustained growth and survival of leukaemic blasts in the bone marrow and periphery, even in the presence of cytotoxic or precision therapies.
Over the last few years, work from my laboratory has attempted to survey all available mitogenic and oncoproteomic contributors to growth, proliferation and treatment resistance in AML blasts (and representative cell line models) utilising a high-resolution, quantitative phosphoproteomic approach. Following training in cell biology3-6 and subsequent optimisation of quantitative proteomic workflows using cancer7, 8 and non-cancer9, 10 systems, I was struck by the number of actionable therapeutic targets revealed via phosphoproteomics – targets that remain invisible via genomics-based approaches.11-13
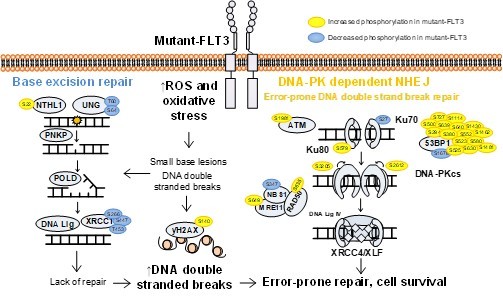
Indeed, our recently published phosphoproteomic analysis of AML patient blasts employed a multimodal analysis pipeline including kinase substrate enrichment analysis,14 coupled with pathway analysis and identified that mutant-FLT3 blasts (biopsied from the bone marrow of de novo AML patients), showing increased phosphorylation of proteins involved in the error-prone DNA-PK-dependent Non Homologous End Joining (NHEJ) pathway, compared to wildtype-FLT3 patients. We examined the preclinical potential of targeting DNA-PK (using M3814/peposertib), and revealed mutant-FLT3 AML blasts were more sensitive than wildtype-FLT3 blasts to DNA-PK inhibition, and when combined with FLT3 inhibition, led to a significant survival advantage both ex vivo and in vivo compared to samples/mice receiving sorafenib alone (p=0.0039).11 This is an exciting treatment paradigm that we hope to move forward into the clinic as a maintenance regimen or at disease recurrence.

Workflow used to identify drug targets invisible to genomics approaches – phosphoproteomic profiling of AML blasts.
So, is phosphoproteomics profiling ready to enter the clinic? Clinical phosphoproteomic profiling has enormous potential for the identification of treatment targets or to be used as an indicator of prognosis, in de novo and refractory settings, in real time, or even as a modality to monitor minimal residual disease. However, we need to consider the methods of patient sample collection, as the phosphoproteome of leukaemic blasts isolated from the bone marrow, will differ from that obtained from peripheral blood; indeed sampling from each simultaneously would reveal the considerable differences in oncogenic signalling influenced by the contrasting physiological environments e.g. oxygen tension, abundance of secreted growth factors, interactions with the stroma, etc. Additionally, the steps taken to enrich leukaemic blasts following bone marrow trephine biopsy or phlebotomy are to be considered; alterations in signalling pathway activity can be influenced by culture media or even the type of blood tube used for collection, and therefore optimised and standardised workflows are required.
The methods used to enrich phosphopeptides and the techniques used to simplify the phosphoproteome to facilitate optimal coverage vary considerably and play a large role in the resolution of the phosphoproteome generated. We must also scrutinise our means of determining statistical significance particularly when using limited numbers of genetically heterogenous patient samples. Corrections for multiple comparisons and conventional p-value cut-offs often fail to identify true positives even when many exist, exacerbated by compression of ratios using tag based quantitative tools.15 A more useful strategy for identification of the biological meaning of proteomics-based treatment targets or biomarkers, is to use false discovery rates coupled with fold-change cut-offs, or to assess the number of key phosphorylation events identified in a pathway from which the novel drug target or biomarker is identified at a global level.
In doing so we will see the advances made in the development of precision therapies thanks to the genomics revolution, augmented by the coupling of proteomic and phosphoproteomic approaches.16 This multi-omics approach may see an AML diagnosis evolve from a death sentence into a manageable condition, similarly to what has been achieved in chronic myeloid leukaemia (CML).
References
- Staudt D, Murray HC, McLachlan T, Alvaro F, Enjeti AK, Verrills NM, et al. Targeting Oncogenic Signaling in Mutant FLT3 Acute Myeloid Leukemia: The Path to Least Resistance. Int J Mol Sci 2018 Oct 16; 19(10).
- Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017 Jan 26; 129(4): 424-447.
- Toop HD, Dun MD, Ross BK, Flanagan HM, Verrills NM, Morris JC. Development of novel PP2A activators for use in the treatment of acute myeloid leukaemia. Org Biomol Chem 2016 May 18; 14(20): 4605-4616.
- Smith AM, Dun MD, Lee EM, Harrison C, Kahl R, Flanagan H, et al. Activation of protein phosphatase 2A in FLT3+ acute myeloid leukemia cells enhances the cytotoxicity of FLT3 tyrosine kinase inhibitors. Oncotarget 2016 Jul 26; 7(30): 47465-47478.
- Dun MD, Anderson AL, Bromfield EG, Asquith KL, Emmett B, McLaughlin EA, et al. Investigation of the expression and functional significance of the novel mouse sperm protein, a disintegrin and metalloprotease with thrombospondin type 1 motifs number 10 (ADAMTS10). Int J Androl 2012 Aug; 35(4): 572-589.
- Dun MD, Smith ND, Baker MA, Lin M, Aitken RJ, Nixon B. The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm-oocyte interaction. J Biol Chem 2011 Oct 21; 286(42): 36875-36887.
- Dun MD, Chalkley RJ, Faulkner S, Keene S, Avery-Kiejda KA, Scott RJ, et al. Proteotranscriptomic Profiling of 231-BR Breast Cancer Cells: Identification of Potential Biomarkers and Therapeutic Targets for Brain Metastasis. Mol Cell Proteomics 2015 Sep; 14(9): 2316-2330.
- Jamaluddin MFB, Ko YA, Kumar M, Brown Y, Bajwa P, Nagendra PB, et al. Proteomic Profiling of Human Uterine Fibroids Reveals Upregulation of the Extracellular Matrix Protein Periostin. Endocrinology 2018 Feb 1; 159(2): 1106-1118.
- Nixon B, De Iuliis GN, Hart HM, Zhou W, Mathe A, Bernstein IR, et al. Proteomic Profiling of Mouse Epididymosomes Reveals their Contributions to Post-testicular Sperm Maturation. Mol Cell Proteomics 2019 Mar 15; 18(Suppl 1): S91-S108.
- Nixon B, Johnston SD, Skerrett-Byrne DA, Anderson AL, Stanger SJ, Bromfield EG, et al. Modification of Crocodile Spermatozoa Refutes the Tenet That Post-testicular Sperm Maturation Is Restricted To Mammals. Mol Cell Proteomics 2019 Mar 15; 18(Suppl 1): S59-S76.
- Murray HC, Enjeti AK, Kahl RGS, Flanagan HM, Sillar J, Skerrett-Byrne DA, et al. Quantitative phosphoproteomics uncovers synergy between DNA-PK and FLT3 inhibitors in acute myeloid leukaemia. Leukemia 2020.
- Dun MD, Mannan A, Rigby CJ, Butler S, Toop HD, Beck D, et al. Shwachman-Bodian-Diamond syndrome (SBDS) protein is a direct inhibitor of protein phosphatase 2A (PP2A) activity and overexpressed in acute myeloid leukaemia. Leukemia 2020 Apr 8.
- Degryse S, de Bock CE, Demeyer S, Govaerts I, Bornschein S, Verbeke D, et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 2018 Mar; 32(3): 788-800.
- Weidner C, Fischer C, Sauer S. PHOXTRACK-a tool for interpreting comprehensive datasets of post-translational modifications of proteins. Bioinformatics 2014 Dec 1; 30(23): 3410-3411.
- Pascovici D, Handler DCL, Wu JX, Haynes PA. Multiple testing corrections in quantitative proteomics: A useful but blunt tool. Proteomics 2016 Sep; 16(18): 2448-2453.
- Murray HC, Dun MD, Verrills NM. Harnessing the power of proteomics for identification of oncogenic, druggable signalling pathways in cancer. Expert Opin Drug Discov 2017 May; 12(5): 431-447.







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in