By: Adam Isabella & Jay Sarthy
The story of cancer is, largely, one of broken genes. The valiant tumor suppressor, defender of cellular integrity, maimed and enfeebled. The conniving oncogene, hyper-activated, allowed to run wild and sow chaos within the cell. These genes are many, and they act on an array of cellular processes. Cell growth. Cell death. Cell motility. Genome integrity. Impairment of this last process is particularly frightening in that it can, in one fell swoop, disrupt a whole suite of cancer-related genes throughout the genome, and ensure that many more will fall in quick succession. Among the defenders of genome integrity are the histones – small protein complexes that wrap DNA strands into orderly bundles. The resulting DNA-histone structure, collectively called chromatin, regulates DNA stability and gene expression. It is not surprising, therefore, that histone function is highly regulated, nor that mutated histones (called oncohistones) with disrupted DNA wrapping characteristics are found in many cancers. There is one type of histone protein, though, that in its native state appears ready-made to cause cancer. Our recent paper in Nature Communications - a collaborative effort between the labs of Drs. Steve Henikoff, Harmit Malik, Robert Bradley, and Marie Bleakley at the Fred Hutchinson Cancer Research Center, led by Dr. Guo-Liang “Chewie” Chew in conjunction with Drs. Antoine Molaro and Jay Sarthy – revealed the role of this unusual histone variant in cancer.
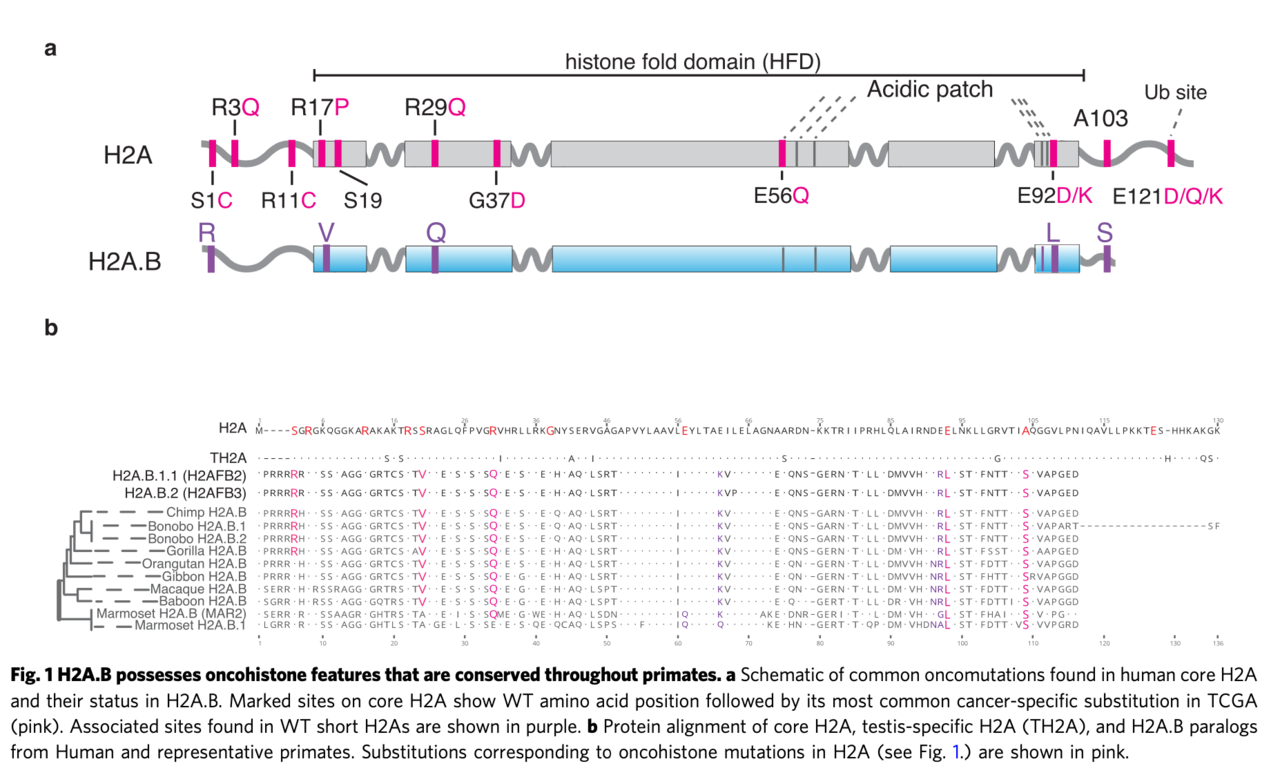
Histones normally wrap DNA tightly and stably around themselves into structures called nucleosomes. Mutations that destabilize nucleosomes are disruptive to genome integrity and, therefore, oncogenic. While most histones are composed of a common set of proteins – called H2A, H2B, H3, and H4 – the mammalian genome contains another class of histones, called the short H2A variants (short H2As). Normally expressed in sperm, short H2As possess a unique characteristic – they destabilize nucleosomes. In other words, exactly what you seemingly don’t want a histone to do. While this doesn’t appear to be a problem for the sperm (more on that below), we hypothesized that these histones would likely be quite problematic if they showed up in another cell type. In fact, by looking more closely at the sequences of the short H2A genes, we made an astonishing discovery: many of the most common cancer-associated mutations in canonical H2A are already present in all wild-type short H2A sequences. Surprisingly, though, these genes had not been identified as cancer-causing agents.
To determine whether short H2As do, as we suspected, contribute to cancer, we examined cancer gene expression databases and found that one class of these genes, H2A.Bs, are indeed frequently turned on in many cancers, including half of diffuse large B-cell lymphomas as well as a smaller number of endometrial, bladder, and cervical carcinomas. Interestingly, while carcinomas were previously shown to have histone mutations, lymphomas were not, suggesting H2A.B misexpression may be another way for cancers to acquire unstable nucleosomes. The cause of activation of H2A.B in these cancers was unclear, although it tended to be co-activated with several other testis-related genes. The consequence of its activation was more evident. H2A.B has previously been observed to interact with splicing factors. Examining H2A.B-reactivated tumors, we uncovered thousands of altered splicing events, with reductions in alternative exon and alternative polyadenylation usage. This genome-wide splicing dysregulation could very well contribute to carcinogenesis, although it is still unclear which particular genes that are dysregulated by H2A.B contribute to cancer.
Oncohistones remain a relatively poorly understood cancer driver. Our work suggests a need to focus not only on mutations to the canonical histone genes, but also on the activities of short H2As as well as other histone variants. Much remains to be explored, including why so many lymphomas reactivate H2A.B, how H2A.B promotes cancer, and whether we can take advantage of H2A.B-driven gene/splicing dysregulation to treat such cancer patients.
Short H2A histones represent a clear and present danger: ready-made oncohistones with a natural ability to destabilize nucleosomes, lurking in our genomes, a ticking time-bomb waiting to be released in the wrong place and wreak havoc on chromatin.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in